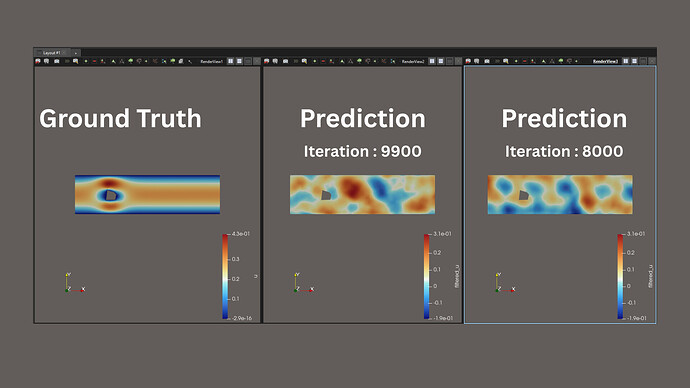
Hello,
I’m currently working on the stokes_mgn example from the NVIDIA PhysicsNemo CFD repository. I was able to successfully train the model as described in the README and also perform physics-informed fine-tuning.
However, I noticed that after fine-tuning, the example code does not save the fine-tuned model. To address this, I made modifications to save the model using both the standard PyTorch save method as well as the PhysicsNemo/Modulus save_checkpoint utility.
I also wrote a separate inference script to run predictions on different .vtp files using this saved fine-tuned model. However, the predictions I get are not as accurate as expected — they are significantly worse than those seen during or after fine-tuning.
These are the modifications I made in code of pi_fine_tuning_gnn.py
after finetuning gets completed I save the model in two ways
logger.info("Physics-informed fine-tuning training completed!")
# Save the model state
model_save_path = os.path.join(to_absolute_path(cfg.results_dir), "pi_fine_tuner.pt")
torch.save(pi_fine_tuner.model.state_dict(), model_save_path)
logger.info(f"Model saved at {model_save_path}")
save_checkpoint(
to_absolute_path(cfg.results_dir),
models=pi_fine_tuner.model,
optimizer=pi_fine_tuner.optimizer,
scheduler=pi_fine_tuner.scheduler,
epoch=cfg.pi_iters,
)
These are the files I created for inferencing
1.For PyTorch method saved model
import os
import numpy as np
import torch
import pyvista as pv
from omegaconf import OmegaConf
from physicsnemo.models.meshgraphnet import MeshGraphNet
from utils import get_dataset
def simple_inference(vtp_file_path, model_path, config_path, output_path=None):
"""
Simple inference function that takes a VTP file and runs the fine-tuned model
Parameters:
-----------
vtp_file_path : str
Path to input VTP file
model_path : str
Path to fine-tuned model (.pt file)
config_path : str
Path to configuration file
output_path : str, optional
Path to save output file
"""
# Set device
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"Using device: {device}")
# Load configuration
cfg = OmegaConf.load(config_path)
# Load the fine-tuned model
print("Loading fine-tuned model...")
model = MeshGraphNet(
cfg.input_dim_nodes + 128, # additional 128 node features from fourier features
cfg.input_dim_edges,
cfg.output_dim,
aggregation=cfg.aggregation,
hidden_dim_node_encoder=cfg.hidden_dim_node_encoder,
hidden_dim_edge_encoder=cfg.hidden_dim_edge_encoder,
hidden_dim_node_decoder=cfg.hidden_dim_node_decoder,
).to(device)
model.load_state_dict(torch.load(model_path, map_location=device))
model.eval()
# Process VTP file
print("Processing VTP file...")
try:
(
ref_u, ref_v, ref_p,
gnn_u, gnn_v, gnn_p,
coords, inflow_coords, outflow_coords, wall_coords, polygon_coords,
nu, dgl_graph
) = get_dataset(vtp_file_path, return_graph=True)
print(f"Successfully processed VTP file with {dgl_graph.number_of_nodes()} nodes")
print(f"Viscosity (nu): {nu}")
except Exception as e:
print(f"Error processing VTP file: {e}")
return None
# Run inference
print("Running inference...")
dgl_graph = dgl_graph.to(device)
with torch.no_grad():
output = model(dgl_graph.ndata["x"], dgl_graph.edata["x"], dgl_graph)
pred_u = output[:, 0:1].detach().cpu().numpy()
pred_v = output[:, 1:2].detach().cpu().numpy()
pred_p = output[:, 2:3].detach().cpu().numpy()
# Calculate errors
def relative_l2_error(pred, ref):
return np.linalg.norm(pred - ref) / np.linalg.norm(ref) * 100
print("\nResults:")
print("Original GNN vs Reference:")
print(f" U error: {relative_l2_error(gnn_u, ref_u):.3f}%")
print(f" V error: {relative_l2_error(gnn_v, ref_v):.3f}%")
print(f" P error: {relative_l2_error(gnn_p, ref_p):.3f}%")
print("\nFine-tuned Model vs Reference:")
print(f" U error: {relative_l2_error(pred_u, ref_u):.3f}%")
print(f" V error: {relative_l2_error(pred_v, ref_v):.3f}%")
print(f" P error: {relative_l2_error(pred_p, ref_p):.3f}%")
# Save results
if output_path is None:
base_name = os.path.splitext(vtp_file_path)[0]
output_path = f"{base_name}_fine_tuned.vtp"
print(f"\nSaving results to: {output_path}")
polydata = pv.read(vtp_file_path)
polydata["fine_tuned_u"] = pred_u.flatten()
polydata["fine_tuned_v"] = pred_v.flatten()
polydata["fine_tuned_p"] = pred_p.flatten()
polydata.save(output_path)
print("Inference completed successfully!")
return pred_u, pred_v, pred_p
# Example usage
if __name__ == "__main__":
# Update these paths according to your setup
vtp_file = "path/to/your/input.vtp" # Your input VTP file
model_file = "path/to/your/pi_fine_tuner.pt" # Your fine-tuned model
config_file = "path/to/your/config.yaml" # Your configuration file
# Run inference
results = simple_inference(vtp_file, model_file, config_file)
- For checkpoint saved method :
import os
import numpy as np
import torch
import pyvista as pv
import dgl
from physicsnemo.models.meshgraphnet import MeshGraphNet
from physicsnemo.launch.utils import load_checkpoint
from hydra.utils import to_absolute_path
from utils import get_dataset
def load_fine_tuned_model(results_dir, device):
"""Load the fine-tuned model with hardcoded config values"""
# Hardcoded config values from config.yaml
cfg = {
"input_dim_nodes": 7,
"input_dim_edges": 3,
"output_dim": 3,
"hidden_dim_node_encoder": 256,
"hidden_dim_edge_encoder": 256,
"hidden_dim_node_decoder": 256,
"aggregation": "sum"
}
model = MeshGraphNet(
cfg["input_dim_nodes"] + 128, # +128 due to Fourier features
cfg["input_dim_edges"],
cfg["output_dim"],
aggregation=cfg["aggregation"],
hidden_dim_node_encoder=cfg["hidden_dim_node_encoder"],
hidden_dim_edge_encoder=cfg["hidden_dim_edge_encoder"],
hidden_dim_node_decoder=cfg["hidden_dim_node_decoder"],
).to(device)
# Load model checkpoint
_ = load_checkpoint(
to_absolute_path(results_dir),
models=model,
device=device,
)
model.eval()
return model
def process_vtp_file(vtp_file_path):
try:
(
ref_u,
ref_v,
ref_p,
gnn_u,
gnn_v,
gnn_p,
coords,
inflow_coords,
outflow_coords,
wall_coords,
polygon_coords,
nu,
dgl_graph,
) = get_dataset(vtp_file_path, return_graph=True)
return dgl_graph, coords, nu
except Exception as e:
print(f"Error processing VTP file: {e}")
raise
def run_inference(model, dgl_graph, device):
dgl_graph = dgl_graph.to(device)
with torch.no_grad():
output = model(dgl_graph.ndata["x"], dgl_graph.edata["x"], dgl_graph)
pred_u = output[:, 0:1].detach().cpu().numpy()
pred_v = output[:, 1:2].detach().cpu().numpy()
pred_p = output[:, 2:3].detach().cpu().numpy()
return pred_u, pred_v, pred_p
def save_results(input_vtp_path, output_path, pred_u, pred_v, pred_p):
polydata = pv.read(input_vtp_path)
polydata["fine_tuned_u"] = pred_u.flatten()
polydata["fine_tuned_v"] = pred_v.flatten()
polydata["fine_tuned_p"] = pred_p.flatten()
polydata.save(output_path)
print(f"Results saved to: {output_path}")
def compare_results(input_vtp_path, pred_u, pred_v, pred_p):
polydata = pv.read(input_vtp_path)
ref_u = polydata.point_data["u"].reshape(-1, 1)
ref_v = polydata.point_data["v"].reshape(-1, 1)
ref_p = polydata.point_data["p"].reshape(-1, 1)
gnn_u = polydata.point_data["pred_u"].reshape(-1, 1)
gnn_v = polydata.point_data["pred_v"].reshape(-1, 1)
gnn_p = polydata.point_data["pred_p"].reshape(-1, 1)
def relative_l2_error(pred, ref):
return np.linalg.norm(pred - ref) / np.linalg.norm(ref) * 100
print("\nOriginal GNN vs Reference:")
print(f" U error: {relative_l2_error(gnn_u, ref_u):.3f}%")
print(f" V error: {relative_l2_error(gnn_v, ref_v):.3f}%")
print(f" P error: {relative_l2_error(gnn_p, ref_p):.3f}%")
print("\nFine-tuned Model vs Reference:")
print(f" U error: {relative_l2_error(pred_u, ref_u):.3f}%")
print(f" V error: {relative_l2_error(pred_v, ref_v):.3f}%")
print(f" P error: {relative_l2_error(pred_p, ref_p):.3f}%")
print("\nFine-tuned vs Original GNN:")
print(f" U difference: {relative_l2_error(pred_u, gnn_u):.3f}%")
print(f" V difference: {relative_l2_error(pred_v, gnn_v):.3f}%")
print(f" P difference: {relative_l2_error(pred_p, gnn_p):.3f}%")
def main():
# Hardcoded inputs
vtp_file = "graph_5.vtp"
results_dir = "finetunecheckpoints"
output_path = vtp_file.replace(".vtp", "_fine_tuned.vtp")
compare = True
# Set device
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"Using device: {device}")
print("Loading fine-tuned model...")
model = load_fine_tuned_model(results_dir, device)
print("Processing VTP file...")
try:
dgl_graph, coords, nu = process_vtp_file(vtp_file)
print(f"Graph has {dgl_graph.number_of_nodes()} nodes and {dgl_graph.number_of_edges()} edges")
print(f"Viscosity (nu): {nu}")
except Exception as e:
print(f"Failed to process VTP file: {e}")
return
print("Running inference...")
pred_u, pred_v, pred_p = run_inference(model, dgl_graph, device)
print("Saving results...")
save_results(vtp_file, output_path, pred_u, pred_v, pred_p)
if compare:
print("Comparing results...")
compare_results(vtp_file, pred_u, pred_v, pred_p)
print("\n✅ Inference completed successfully!")
print(f"👉 Fine-tuned predictions saved in: {output_path}")
if __name__ == "__main__":
main()
Questions:
- What is the correct way to save and load a fine-tuned model in the PhysicsNemo framework to preserve all necessary state for inference?
- What is the role of the
.mdlusfile generated during checkpoint saving? Should it be used during inference, and how? - Could someone take a look at the modifications I made to save the model and my inference script, and suggest improvements or corrections?
Thanks for support !!