Hello all,
I am really hoping to sell my boss on the idea of CUDA. I have a ridiculously stubborn Research Adviser that would rather spend thousands on racks of old dual socket Barcelona U2 than take a chance on CUDA (even outside of our cluster :shrug:)
So I decided to venture a bit on my own. I was fortunate enough to attend a ‘Many Cores’ Seminar at the Ohio Supercomputer center last year and my cuda programming is slowing developing, but the only place my boss would care to see improvement is on our NAMD bio system jobs.
My desktop rig is not that hot for crunching numbers. I would prefer some more cores (maybe I’ll toss in a q9650 :shrug:)
E8600 @ 3.96Ghz (this is a dual core wolfdale with 6mb of l2)
GTX 260 (192sp cards) @ stock 576mhz core
CentOS 5.5
So I tossed on NAMD 2.7 as well as the NAMD 2.7 CUDA version and am running two separate systems.
System 1: Water box (50 x 50 x 50) in which I am growing a micelle from Perfluorooctanoate. The system is neutralized with sodium. Please ignore the periodic boundary caused flip flopping. The box is too small for this one, but it was a nice test npt simulation. (This was my undergrad senior project, I just graduated :) )
Here I am just representing the surfactant resi’s
It’s fun to watch them ‘bud’ from the water voids in the .dcd’s
System 2: I have a lipid bilayer than I am playing with. This system has many more atoms, so I was hoping to see some better CUDA benefits to show the boss.
Parameters:
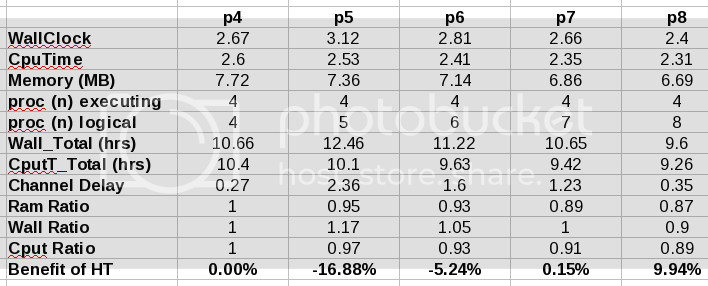
Results:
My WallTimes were cut in HALF for my micelle system. Considering all of the bonded interactions are still only taking place on my dual core (albeit @ 3.9Ghz) I was very happy to see this. I am still waiting for my cpu only run of system 2. Extrapolating from past data, it looks like it may be about a 130% speed up.
Conclusions/Remarks: I am really trying to sell my boss on the idea of putting GTX470’s in a couple of the cluster machines or some of the desktops for submitting these type of jobs too. And I was hoping to get some ammunition from you guys. Is there anything obvious that I am missing?
I noticed that only one of my two GTX260’s is being used for the non-bonded calculations and it barely heats up at all. Is there any way I can unload more work on the GPU?
PS. I submit with the following command lines:
I cannot seem to find any options that would allow me to tweak how my card is being used here. I have grown to hate the NAMD manuals…
Thank you for your time,
Luke